Mapas auto-organizativos. Self self organizing map (SOM)¶
# pip install susi
Carga de librerías necesarias¶
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import susi
from susi.SOMPlots import plot_nbh_dist_weight_matrix, plot_umatrix, plot_estimation_map
Carga de datos. Datos vitivinícolas¶
ColNames = ["Cultivars","Alcohol","Malic_acid","Ash","Alcalinity_of_ash",
"Magnesium","Total_phenols","Flavanoids","Nonflavanoid_phenols",
"Proanthocyanins","Color intensity","Hue","OD280/OD315","Proline"]
df = pd.read_csv('./data/wine.csv', header=None, names=ColNames)
df.head()
| Cultivars | Alcohol | Malic_acid | Ash | Alcalinity_of_ash | Magnesium | Total_phenols | Flavanoids | Nonflavanoid_phenols | Proanthocyanins | Color intensity | Hue | OD280/OD315 | Proline | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 14.23 | 1.71 | 2.43 | 15.6 | 127 | 2.80 | 3.06 | 0.28 | 2.29 | 5.64 | 1.04 | 3.92 | 1065 |
| 1 | 1 | 13.20 | 1.78 | 2.14 | 11.2 | 100 | 2.65 | 2.76 | 0.26 | 1.28 | 4.38 | 1.05 | 3.40 | 1050 |
| 2 | 1 | 13.16 | 2.36 | 2.67 | 18.6 | 101 | 2.80 | 3.24 | 0.30 | 2.81 | 5.68 | 1.03 | 3.17 | 1185 |
| 3 | 1 | 14.37 | 1.95 | 2.50 | 16.8 | 113 | 3.85 | 3.49 | 0.24 | 2.18 | 7.80 | 0.86 | 3.45 | 1480 |
| 4 | 1 | 13.24 | 2.59 | 2.87 | 21.0 | 118 | 2.80 | 2.69 | 0.39 | 1.82 | 4.32 | 1.04 | 2.93 | 735 |
Se estándariza el conjunto de datos de entrada¶
Para lo que se hace uso de la librería StandardScaler de sklearn. Si además hubiese algún atributo etiquetado sin valores continuos previamente a la estandarización se etiquetarían estas columnas del dataset haciendo uso de la librería LabelEncoder de sklearn.preprocessing.
from sklearn.preprocessing import StandardScaler
sc = StandardScaler()
df[["Alcohol","Malic_acid","Ash","Alcalinity_of_ash", "Magnesium","Total_phenols","Flavanoids","Nonflavanoid_phenols",
"Proanthocyanins","Color intensity","Hue","OD280/OD315","Proline"]] = sc.fit_transform(df[["Alcohol",
"Malic_acid","Ash","Alcalinity_of_ash", "Magnesium","Total_phenols","Flavanoids",
"Nonflavanoid_phenols", "Proanthocyanins","Color intensity","Hue","OD280/OD315","Proline"]])
df.head()
| Cultivars | Alcohol | Malic_acid | Ash | Alcalinity_of_ash | Magnesium | Total_phenols | Flavanoids | Nonflavanoid_phenols | Proanthocyanins | Color intensity | Hue | OD280/OD315 | Proline | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 1.518613 | -0.562250 | 0.232053 | -1.169593 | 1.913905 | 0.808997 | 1.034819 | -0.659563 | 1.224884 | 0.251717 | 0.362177 | 1.847920 | 1.013009 |
| 1 | 1 | 0.246290 | -0.499413 | -0.827996 | -2.490847 | 0.018145 | 0.568648 | 0.733629 | -0.820719 | -0.544721 | -0.293321 | 0.406051 | 1.113449 | 0.965242 |
| 2 | 1 | 0.196879 | 0.021231 | 1.109334 | -0.268738 | 0.088358 | 0.808997 | 1.215533 | -0.498407 | 2.135968 | 0.269020 | 0.318304 | 0.788587 | 1.395148 |
| 3 | 1 | 1.691550 | -0.346811 | 0.487926 | -0.809251 | 0.930918 | 2.491446 | 1.466525 | -0.981875 | 1.032155 | 1.186068 | -0.427544 | 1.184071 | 2.334574 |
| 4 | 1 | 0.295700 | 0.227694 | 1.840403 | 0.451946 | 1.281985 | 0.808997 | 0.663351 | 0.226796 | 0.401404 | -0.319276 | 0.362177 | 0.449601 | -0.037874 |
X = df.values[:,1:14]
y = df.values[:,0]
X.shape, y.shape
((178, 13), (178,))
Se clasifica¶
som = susi.SOMClassifier(
n_rows=25,
n_columns=25,
n_iter_unsupervised=1000,
n_iter_supervised=1000,
random_state=0)
som.fit(X, y)
y_pred = som.predict(X)
print("Accuracy: {0:.1f} %".format(som.score(X, y)*100))
Accuracy: 91.0 %
Se imprime la matriz U¶
u_matrix = som.get_u_matrix()
plot_umatrix(u_matrix, 25, 25)
plt.show()

Se imprime la matriz de vecindad¶
plot_nbh_dist_weight_matrix(som)
plt.show()
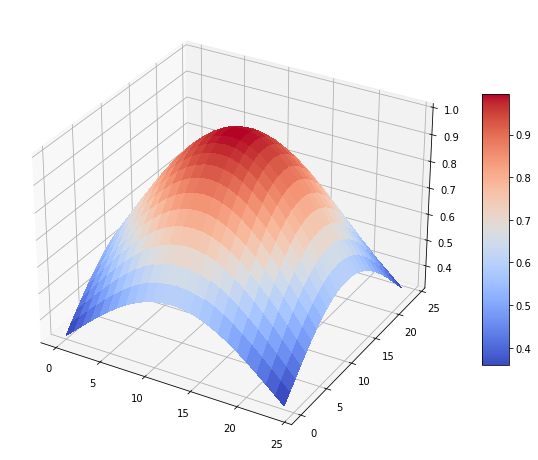
Se imprime el mapa de estimaciones¶
estimation_map = som.get_estimation_map().squeeze()
plot_estimation_map(estimation_map)
plt.show()

